I've been learning a lot about PostgreSQL recently. A trigger for this was a Stack overflow question I came about some time ago, featuring a fairly complex equation for calculating color similarity. I've been obsessing over it ever since and I've used it as a test case for a number of things. Hopefully, this will be one of several articles about how I used it and what I learned along.
This article is a benchmark of different implementations of same function in PostgreSQL. The function in question is difference between colors, and the implementations are in SQL, C, and several variants of Python: pure, numpy, numba and Cython.
Delta E color difference is a measure of color difference. The L*a*b color space was designed with intention to represent colors in a perceptually uniform way, meaning some change in value should result in equal or very similar change in perception. Since the color space was designed with this intention, the original CIE 1976 Delta E was just cartesian distance between two point in color space. Along with more research it became clear the CIE 1976 had some deficiencies and was followed by improved version CIE 1994 Delta E. The latest revision is CIE 2000 Delta E and is far from being a simple equation. For the reader to understand the rest of the post, it is enough to know this equations is math intensive, accepts two 3-dimensional vectors, and returns a single scalar value.
So, with the CIE 2000 Delta E standard no longer being a simple calculation, implementing it in SQL became cumbersome. Jaza, author of the SO question, managed to implement it in SQL (kudos!), but even with CTEs the code is still incomprehensible. And the code is slow, too. SQL just wasn't made for this.
In an effort to speed this up, I tested several approaches. I tested Jaza's SQL implementation, plpgsql implementation and my custom PostgreSQL C extension (more about this in another post). PostgreSQL officially supports Python as a server side procedural language, so this is also something to evaluate. Python is diverse and offers several options for implementation, depending on your needs for performance: pure Python, numpy, numba, cython.
For testing I used several configurations: pure SQL function (as written by Jaza), plpgsql function, my PostgreSQL C extension, pure Python function, Python function using NumPy, then I used Python module as a container for functions, and additionally tried to see if I can make numba JIT work on the server side.
The results are interesting, but first the setup.
Implementations
SQL, plpgsql and C implementations are just functions and there's not much wiggle room.
There are several Python implementations:
- inline pure
- inline pure jitted
- inline numpy
- inline numpy jitted
- module pure
- module pure jitted
- module numpy
- module numpy jitted
- cython module
"Inline" or "module" defines if the function body is defined inline in CREATE FUNCTION statement or in a separate Python module, which is imported at execution.
"Numpy" or "pure" signifies the functions uses colormath code (which in turn uses numpy) or a pure Python implementation.
"Jitted" signifies the use of numba jitting capabilities.
Setup
I created the test database:
dropdb colors createdb colors psql colors -c 'create table color (rgb_r double precision, rgb_g double precision, rgb_b double precision, lab_l double precision, lab_a double precision, lab_b double precision);'
I then generated random test dataset and used the colormath module to convert the RGB colors to Lab colorspace in order for the values to represent a valid RGB value.
Benchmark
For benchmark I created a table with 20.000 random RGB colors, converted them to Lab color space and stored in database. Test case consists of running the distance function on whole table, comparing with a fixed value, in my case it was L=42.041, a=-9.105, b=-13.873. Each implementation was called 10 times on an idle laptop.
Reusing connections
First run was with connection reuse:
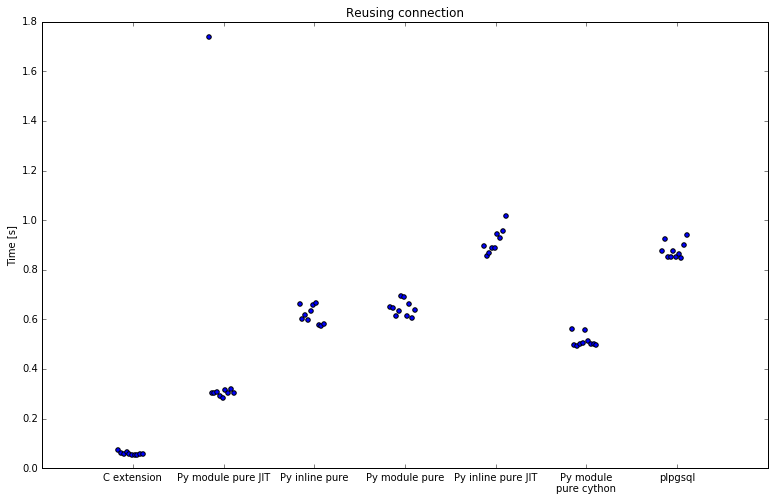
| Implementation | Minimum | Average | Maximum | Speedup |
|---|---|---|---|---|
| SQL | 11.3849 | 11.8725 | 12.5735 | 1.0 |
| plpgsql | 0.8482 | 0.8786 | 0.9386 | 13.5 |
| C extension | 0.0550 | 0.0593 | 0.0715 | 200.3 |
| Py inline pure | 0.5735 | 0.6166 | 0.6651 | 19.3 |
| Py inline pure JIT | 0.8567 | 1.2102 | 3.8561 | 9.8 |
| Py inline pure JIT cached | 0.8730 | 0.9800 | 1.2758 | 12.1 |
| Py inline numpy | 6.2782 | 6.5544 | 6.8891 | 1.8 |
| Py module pure | 0.6055 | 0.6445 | 0.6936 | 18.4 |
| Py module pure JIT | 0.2837 | 0.4468 | 1.7373 | 26.6 |
| Py module pure JIT cached | 0.2878 | 0.4241 | 1.4920 | 28.0 |
| Py module pure cython | 0.4944 | 0.5123 | 0.5600 | 23.2 |
| Py module numpy | 6.0890 | 6.4830 | 7.7312 | 1.8 |
| Py module numpy JIT | 6.0668 | 6.5032 | 7.5097 | 1.8 |
| Py module numpy JIT cached | 6.0912 | 6.5865 | 7.3711 | 1.8 |
SQL is slowest with 11 seconds for 20k rows. C extension is fastest, with 55 to 72 miliseconds and offers a speedup of 200x.
In between there's Python. All numpy implementations are a bit less than 2x faster than SQL. Using numpy in plpython is slow, since the data needs to be copied to numpy array before it is processed. This is a costly operation and it shows. Inline numpy with numba JIT took far longer that SQL against which I am comparing so I didn't test it at all.
Pure Python implementations do better and are about 19x faster than SQL. By defining function inline instead of in module it was 4% faster, but that can make logic harder to maintain. However, when using numba JIT, defining function inline slows down execution to 12x faster, and defining function in module speeds it up to 28x faster. Apparently defining function inline makes JIT less effective. All numba decorated functions also have an initialization penalty, where the first run is significantly slower, most obvious with pure jit module.
Caching with numba does not seem to help. It does have one extra requirement, since the server process needs write permissions on directory next to python module in order to write cached data to disk.
I also tested a Cython implementation, at 23x it was faster than pure Python, but slower than numba.
New connections
Second run was with new connection for every execution:
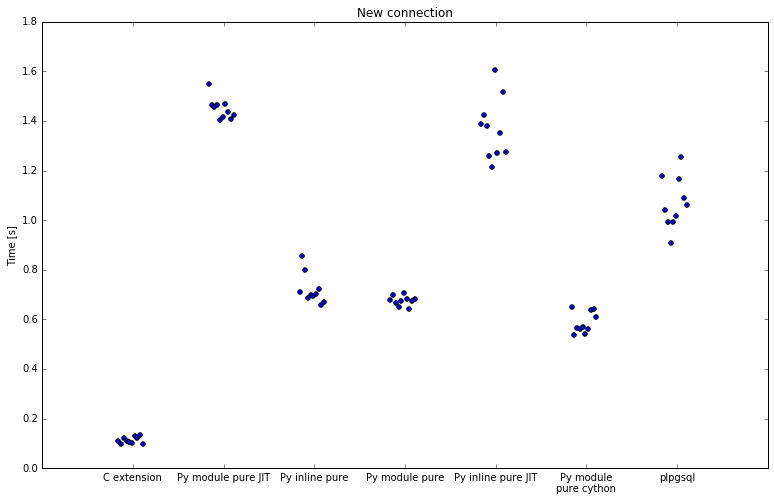
| Implementation | Minimum | Average | Maximum | Speedup |
|---|---|---|---|---|
| SQL | 11.5494 | 12.0963 | 12.7010 | 1.0 |
| plpgsql | 0.9093 | 1.0696 | 1.2528 | 11.3 |
| C extension | 0.0977 | 0.1129 | 0.1325 | 107.2 |
| Py inline pure | 0.6567 | 0.7199 | 0.8560 | 16.8 |
| Py inline pure JIT | 1.2135 | 1.3678 | 1.6060 | 8.8 |
| Py inline pure JIT cached | 1.1845 | 1.2449 | 1.3095 | 9.7 |
| Py inline numpy | 6.0845 | 6.4325 | 6.7866 | 1.9 |
| Py module pure | 0.6414 | 0.6754 | 0.7062 | 17.9 |
| Py module pure JIT | 1.4029 | 1.4491 | 1.5507 | 8.3 |
| Py module pure JIT cached | 1.4275 | 1.5280 | 1.6158 | 7.9 |
| Py module pure cython | 0.5374 | 0.5874 | 0.6513 | 20.6 |
| Py module numpy | 7.1427 | 7.3657 | 7.5970 | 1.6 |
| Py module numpy JIT | 7.2352 | 7.4535 | 7.7606 | 1.6 |
| Py module numpy JIT cached | 7.3986 | 7.5940 | 7.9226 | 1.6 |
When including connection overhead there are significant reductions in performance. It is important to understand how PostgreSQL implements plpython and server processes in general. After connecting, PostgreSQL forks a process for client. If you run any plpython functions, this process then loads Python interpreter, also adding some overhead.
SQL implementation stays almost the same, numpy is slightly slower with 1.6-1.9x speedup.
All implementations with numba are significantly slower. Where numba was previously 28x faster, it is now always hitting initialization on first request, and the speedup drops to 7.9x. Best numba enabled implementation is 9.7x faster, down from 12.1x. Both are slower than pure Python.
A couple of implementations stand out here: pure Python, both inline and as module, and also Cython, have approximately the same speedup as before, meaning they are a stable implementation and do not incur significant initialization overhead.
Lessons learned
Compared by performance we get three clusters of implementations:
- SQL and numpy are in the same order of magnitude (slow),
- plpgsql and pure Python implementations are an order of magnitude faster and
- C function is two orders of magnitude faster.
In plpython, you don't want to use numpy, which is slow because each row is processed individually. You might want to use numba if you're reusing connections, but care should be taken, since it can have unpredictable performance. Cython is okay, though it probably won't help much over pure Python. Functions, written in C, are fast.